Improve the specificity and sensitivity of your assay (PCR, microassay,..) by including Locked Nucleic Acid (LNA) bases! You can vary the number of LNA bases to 'fine-tune' the Tm of a nucleotide duplex. This can ensure discrimination between closely related sequences (detection of mutations or SNPs).
Locked Nucleic Acid is a type of nucleic acid analog that contains a 2'-O, 4'-C methylene bridge.
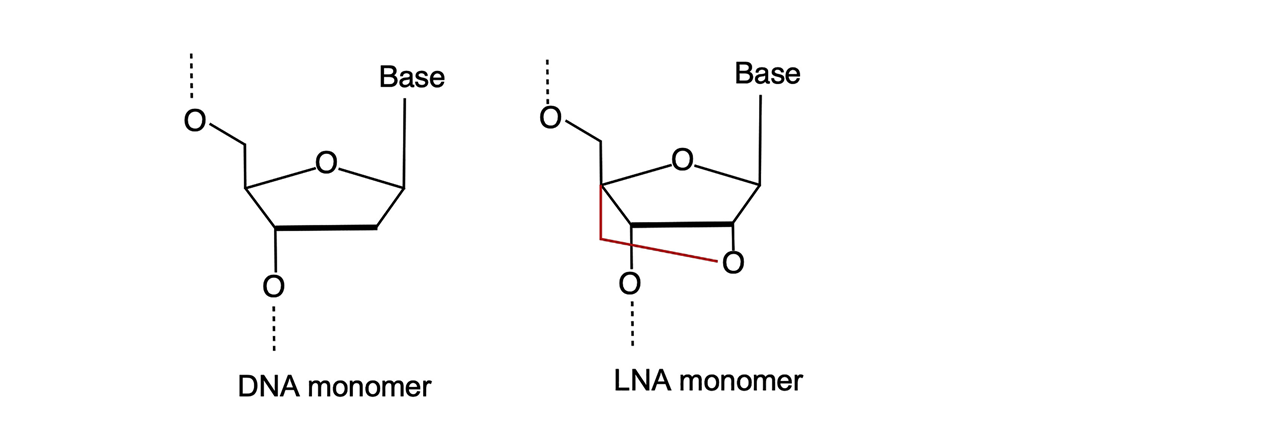
Figure 1. Comparative structure of native-state DNA and Locked Nucleic Acid (LNA) monomers (1).
The “locked” part of their name comes from a methylene bridge bond linking the 2′-oxygen to the 4′-carbon of the RNA pentose ring (Figure 1, red). The bridge bond fixes the pentose ring in the 3′-endo conformation. In fact, LNA nucleotides are RNA building blocks due to the 2´-oxygen on the sugar component in place, however not requiring a 2´OH protection group (like TBDMS, TOM, ACE or TC) as “the lock” provides protection.
LNA building blocks in PCR primers, qPCR probes, and other types of oligonucleotides provide “oligo solubility” in water and standard buffers. And as the modification concerns the Nucleic Acid backbone only, Watson-Crick base-pairing rules remain unaffected.
LNA modified oligonucleotides advantages at a glance
... featuring
- Increased thermal stability and hybridization specificity
- Increased Tm for short and AT-rich primers and probes (ΔTm/locked base approx. 2-6 °C depending on sequence composition and “neighbourhood” effects). While approximations of the Tm-increasing effect of MGB and LNA can be found, an exact calculation of the final Tm is not possible and cannot be provided a priori. The Tm-increasing effect depends strongly on salt concentrations, pH, temperature, sequence composition, GC content, oligo length, and other biophysical/biochemical parameters and reaction conditions; hence, an optimization of each assay must be performed
- Enhanced in vitro and in vivo stability due to increased endo- and exonuclease resistance
- Nontoxic, efficient and effective building blocks for incorporation in antisense, m-RNA or other Nucleic Acid based drug development candidates.
... providing
- Efficient mismatch/SNP/allelic discrimination
- Earlier Ct values
- Higher signal level/signal-to-noise ratio
- Facilitated and flexible designs for problematic target sequences – however sequence dependent
- an excellent supplementation to our Tm/specificity/sensitivity increasing portfolio: ZNA and MGB modified oligonucleotides. Worth keeping in mind that LNAs, as well as ZNA®s and MGBs, exhibit similar properties in terms of increasing affinity for the target (increasing Tm), however, ZNA has unique features, such as enhanced/accelerated target recognition, rendering it feasible to use to reduce the cycling time or enhance the accuracy of quantification of low abundant transcripts.
... hence qualifying for
a wide range of Nucleic Acid-based applications like
- PCR/real-time PCR/RT-PCR
- Microarrays/Capture probing
- Gene silencing
- Fluorescent In-Situ Hybridization (FISH).
Our standard LNA Primers & Probes - Portfolio
5 yield ranges based on independent lengths of delivered oligonucleotide quantities from 5 to 60 base lengths for LNA primers and from 6 to 40 base lengths for LNA probes, at a maximum LNA moiety of 15. Please note that HPLC purification and QC by Mass-Check are included.
- ≥ 5 < 10 nmol
- ≥ 10 < 20 nmol
- ≥ 20 < 30 nmol
- ≥ 30 < 50 nmol
- ≥ 50 < 70 nmol
For ranges ≥ 70 nmol, please inquire. An add-on price is charged for LNA-DLPs blocks in addition to the basic DLPs price.
References
FAQ – you ask, we answer
#
The preferred way is order transmission through our Web Order Portal for most convenient online shopping (please refer here for further details).
Alternatively orders can be placed by sending us an e-mail at oligo@metabion.com with our pre-formatted Excel order file as attachment.
In the section “Dual labelled probes” you can find various 5’ Reporter-3´ Quencer combinations. Select the one of your choice and mark the respective LNA nucleotides in the sequence as (+A), (+C), (+G), and (+T) respectively.
R-Q combinations and yield ranges exceeding our standard portfolio can certainly be inquired.
#
When designing LNA-containing oligonucleotides, one should follow the basic rules of primer design particularly pay attention to the location and number of LNAs.
For example, a typical 18-mer should contain a maximum of 7–8 LNAs.
Also, try to avoid stretches of more than 4 consecutive LNAs, which would result in very tight hybridization in that region.
Stretches of LNAs are to be avoided close to the 3′ end of an oligonucleotide. LNA will bind very tightly to other LNA residues. Avoid self-complementarity and cross-hybridization to other LNA-containing oligonucleotides.
Finally, be sure to match the Tm of the primers, as usual, keeping in mind that each substitution of a standard nucleotide with an LNA increases the Tm by 2-6 °C per LNA.
For novel applications, design guidelines may have to be established empirically by creating your own “best recipe”.
#
Yes, this is possible. Please mark the respective phosphorothioate bond by an asterix (*).
A fully thioated LNA 5´ATCGAT3´ Hexamer for example shall be depicted as follows:
5´ (+A) *(+T) *(+C) *(+G) *(+A) *(+T) 3´
#
For LNA probes selected from our standard portfolio you can expect a TAT of 5 business days.
#
A hardcopy of the preparative HPLC chromatogram and the mass spectrum are included in addition to the respective synthesis report and delivery note.
#
A concentration of 0.2 µM of the LNA probe should be fine for most assays, but as usual the optimal concentration should be determined experimentally. Due to the higher affinity of the probe to the target, the probe concentration might be lower.
#
Locked Nucleic Acids (LNA) probes rely on modified nucleotide chemistry with regard to the sugar component involved, while the organic base component is “unmodified” and thus follows Watson-Crick base-pairing rules when mixed with DNA or RNA bases in an oligonucleotide. When incorporated into an oligonucleotide probe, locked nucleic acid monomers increase structural stability, resulting in a raise of the formed duplex´melting temperature (Tm). Locked Nucleic Acids are not recognized by DNA/RNAses as a substrate, hence LNA modified oligonucleotides also display significant resistance to nucleases.
In contrast, ZNA®s are oligonucleotides conjugated with repeated cationic spermine units that decrease electrostatic repulsions with target nucleic acid strands, and greatly improve hybridization properties by enhanced affinity to the complementary target sequence as well as increased stability of the formed duplex at an unprecedented specificity. The “Tm boost” generated by adding ZNA to either end of the oligonucleotide probe is significant and sequence-independent, meaning the core sequence remains “unmodified”.
Minor Groove Binder (MGB) probes, ZNA®s, and LNAs are known to increase the Tm of an oligo sequence. MGB probes include a minor groove binder moiety at the 3’ end that increases the melting temperature of the probe and stabilizes the hybridization of the probe DNA to its target sequence. The introduction of the minor groove binder moiety is sequence-independent, too.
In summary, metabion offers all three duplex stability enhancing modifications, and therefore provides greatest flexibility in assay design and choosing the right option for your required application.

